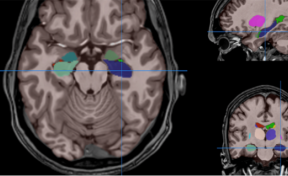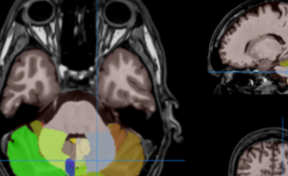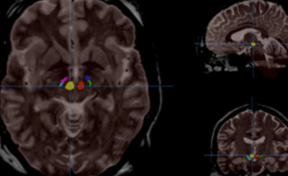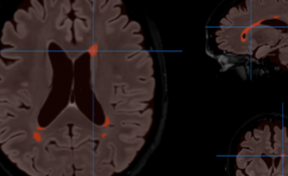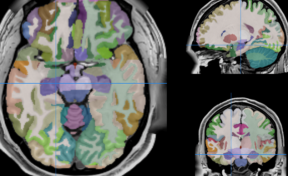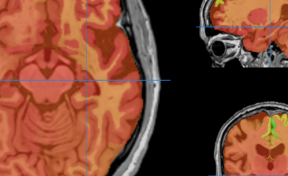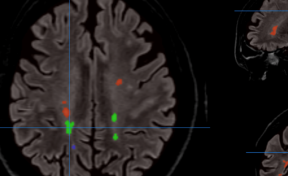
HIPS : Software providing hippocampus subfield segmentation based on two different delineation protocols using both monomodal (T1w) and multimodal (T1w and T2w) MR images
Domaine Santé et Bien-être
Technologies
Technologies d'imagerie pour la santé
Exploitation numérique des données de santé
Challenges
The HIPS module automatically analyzes and provides quantitative analyses of lesions in the hippocampal subfields related to several pathologies (multiple sclerosis, Alzheimer's disease, vascular brain injuries, etc.). This module is particularly efficient for the early detection of Alzheimer's disease and Multiple Sclerosis.
Innovative solution
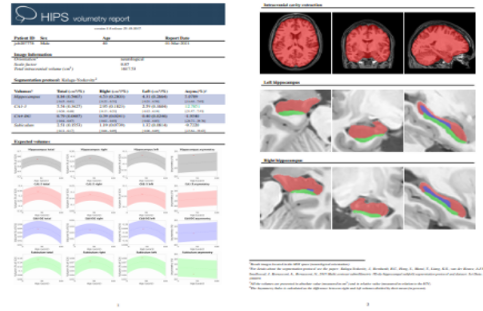
HIPS is a pipeline for automatic hippocampus subfield segmentation for monomodal (T1) or multimodal MRI data (T1 + T2). It gets anonymized MRI brain volumes in NIFTI format and produces a pdf report with the volumes of different subfields using two different delimitation protocols. The average processing time of the whole pipeline is around 20 minutes.
APPLICATIONS
-
Clinical trials
-
Companion diagnostics
-
Clinical routine
COMPETITIVES ADVANTAGES
-
No need of learning complex software packages
-
No need having expensive computational infrastructures in your local site
-
Faster and more accurate than other solutions
-
HIPS works in a fully automatic manner and is able to provide brain structure volumes without any human interaction
-
Two reports (CSV and PDF) providing all the volumetry values calculated from the segmentations as well as asymmetry indexes
How it works
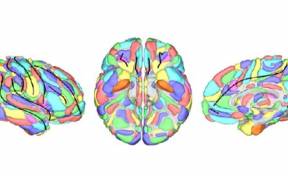
HIPS is a segmentation tool based on a multi-atlas patch-based method, capable of automatically segmenting hippocampal images from T1w and/or T2w MRI.
HIPS is integrated into the volBrain web service. First, the user provides a 3D T1w and/or T2w MRI to the system. The system will first preprocess the image (denoising, inhomogeneity correction, registration) and then segment the hippocampal subfields.
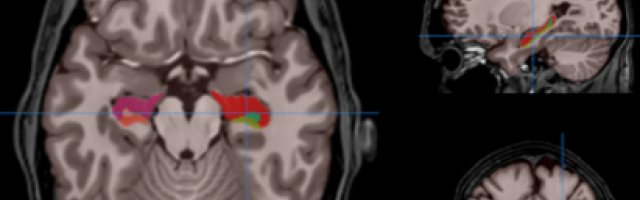
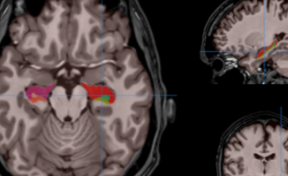
HIPS has two different delineation protocols, Winterburn (5 subfields) or Kulaga-Yoskovitz (3 subfields).
Winterburn protocol labels are: CA1, CA2/3, CA4/DG, SR/SL/SM and Subiculum.
Kulaga-Yoskovitz protocol labels are: CA1-3, CA4/DG and Subiculum.
Once the process is finished, you will be notified by e-mail so you will be able to download a package including some image files and two reports (CSV and PDF) offering all the volumes estimated from the segmentations. The PDF includes patient information, lesion classes, volumes and their locations in MNI space. It also includes several snapshots from the different labeling steps as a quality control.
Inventeurs
Developed by Pierrick COUPÉ (LaBRI - université de Bordeaux, CNRS, Bordeaux INP) & Jose Vicente MANJON HERRERA (Universitat Politècnica de València)
Propriété intellectuelle
Software registered with the Program Protection Agency APP
PARTERSHIPS
License to companies providing :
- Global healthcare solutions
- Diagnostic medical imaging solutions
- Medical imaging equipment
Contact
Jean-Luc CHAGNAUD
%6a%6c%2e%63%68%61%67%6e%61%75%64%40%61%73%74%2d%69%6e%6e%6f%76%61%74%69%6f%6e%73%2e%63%6f%6d
Associated innovations
- VolBrain A software to analyze MRI brain data. It provides a report including volume information on brain structures.
- CERES : A software to automatically analyze the cerebellum in brain MRI to provide a report including volume information on different aspects of the cerebellum
- pBrain : A software providing Parkinson’s disease related deep nucleus segmentation (Substantia Nigra locus niger), Red Nucleus and Subthalamic nucleus) using T2w MR images.
- LesionBrain : A software providing IC extraction, brain tissue classification and delineation/ classification of white matter hyperintense lesions using T1w and FLAIR MR images.
- AssemblyNet : A software providing segmentation of the intracranial cavity, brain tissues (WM, subcortical GM, cortical GM, CSF), cortical lobes, cortical and subcortical structures using a T1w MR image.
More…
HIPS is available on : www.volbrain.net